Introduction
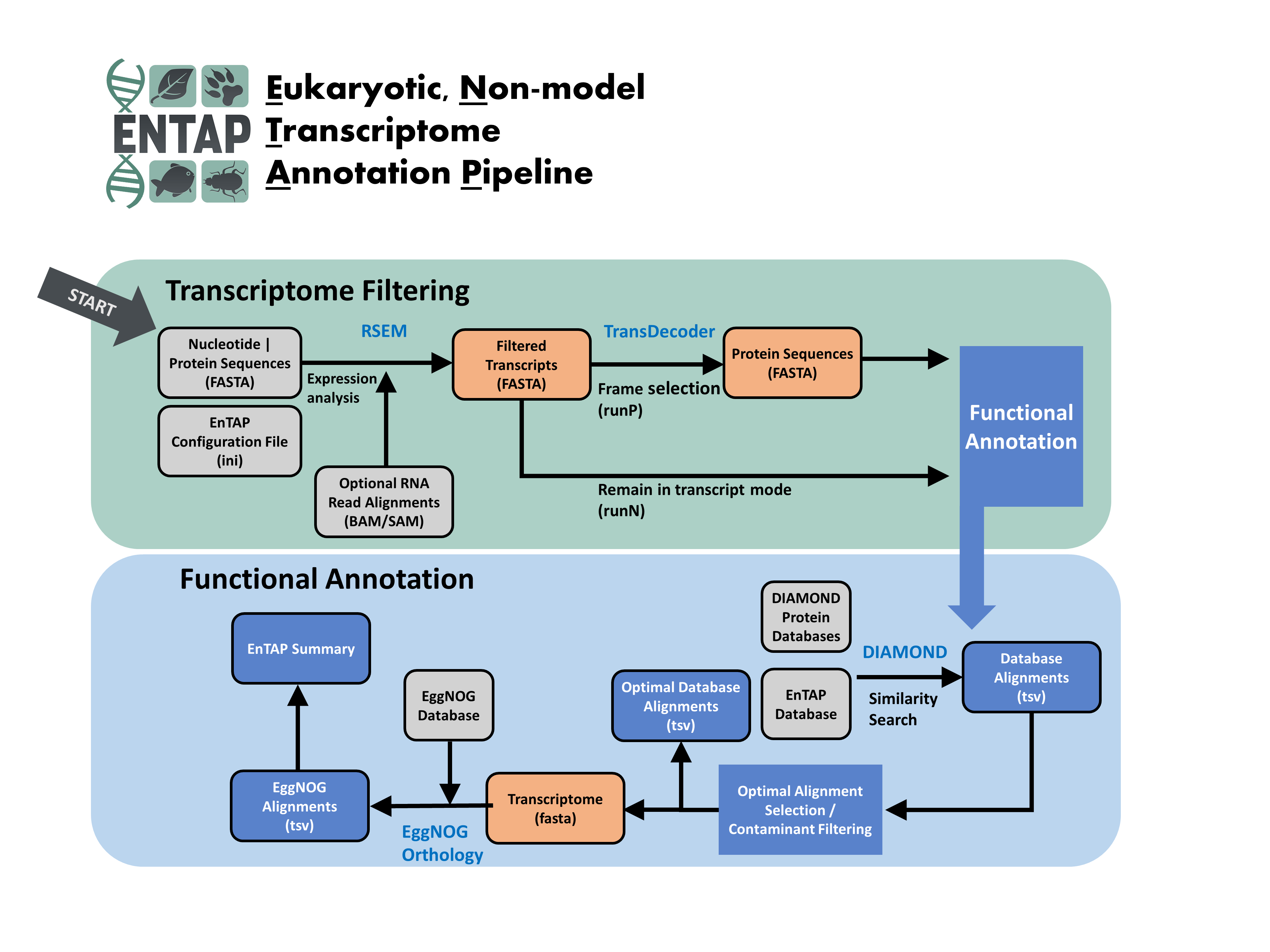
The Eukaryotic Non-Model Transcriptome Annotation Pipeline (EnTAP) is designed to improve the accuracy, speed, and flexibility of functional gene annotation for de novo assembled transcriptomes in non-model eukaryotes.
This software package addresses the fragmentation and related assembly issues that result in inflated transcript estimates and poor annotation rates. Filtering is applied through assessment of true expression and frame selection, followed by leveraging open-source tools to functionally annotate the translated proteins.
Downstream features include fast similarity search across multiple databases, protein domain assignment, orthologous gene family assessment, Gene Ontology term assignment, and KEGG pathway annotation.
The final annotation integrates across multiple databases and selects an optimal assignment from a combination of weighted metrics describing similarity search score, taxonomic relationship, and informativeness. Researchers have the option to include additional filters to identify and remove potential contaminants and prepare the transcripts for enrichment analysis. This fully featured pipeline is easy to install, configure, and runs much faster than comparable functional annotation packages. It is developed to contend with many of the issues in existing software solutions.
Pipeline Stages
- Transcriptome Filtering: designed to remove assembly artifacts and identify true CDS (complete and partial genes)
Expression Filtering (RSEM)
Frame Selection (TransDecoder by default, or GeneMarkS-T)
- Transcriptome Annotation: designed to assign functional information to sequences (homology, Gene Ontology, KEGG)
Similarity Search: optimized search against user-selected databases (DIAMOND).
Contaminant Filtering and Best Hit Selection: selects final annotation and identifies potential contaminants
Orthologous Group Assignment: independent assignment of translated protein sequences to gene families (eggNOG). Includes protein domains (SMART/Pfam), Gene Ontology (GO) terms, and KEGG pathway assignment.
InterProScan (optional): sequence search against the families of InterPro databases to assign protein domains, Gene Ontology terms, and pathway information
- How to cite:
Hart AJ, Ginzburg S, Xu M, et al. EnTAP: Bringing faster and smarter functional annotation to non-model eukaryotic transcriptomes. Mol Ecol Resour. 2020;20:591–604. https://doi.org/10.1111/1755-0998.13106
Citations
- [1] A. Mitchell, H.-Y. Chang, and L. Daugherty, “The InterPro protein families database: the
classification resource after 15 years,” Nucleid Acids Research, vol. 43, no. D1, 2015.
- [2] B. Buchfink, Xie C., D. Huson, “Fast and sensitive protein alignment using
DIAMOND”, Nature Methods 12, 59-60 (2015).
- [3] eggNOG 4.5: a hierarchical orthology framework with improved functional
annotations for eukaryotic, prokaryotic and viral sequences. Jaime Huerta-Cepas, Damian Szklarczyk, Kristoffer Forslund, Helen Cook, Davide Heller, Mathias C. Walter, Thomas Rattei, Daniel R. Mende, Shinichi Sunagawa, Michael Kuhn, Lars Juhl Jensen, Christian von Mering, and Peer Bork. Nucl. Acids Res. (04 January 2016) 44 (D1): D286-D293. doi: 10.1093/nar/gkv1248
- [4] G. O. Consortium, “Gene Ontology Consortium: going forward,” (in eng), Nucleic Acids Res,
vol. 43, no. Database issue, pp. D1049-56, Jan 2015.
- [5] J. Huerta-Cepas et al., “Fast genome-wide functional annotation through orthology
assignment by eggNOG-mapper,” (in eng), Mol Biol Evol, Apr 2017.
- [6] M. Kanehisa, M. Furumichi, M. Tanabe, Y. Sato, and K. Morishima, “KEGG:
new perspectives on genomes, pathways, diseases and drugs,” (in eng), Nucleic Acids Res, vol. 45, no. D1, pp. D353-D361, Jan 2017
- [7] B. Li and C. N. Dewey, “RSEM: accurate transcript quantification from
RNA-Seq data with or without a reference genome,” (in eng), BMC Bioinformatics, vol. 12, p. 323, Aug 2011.
- [8] P. Jones et al., “InterProScan 5: genome-scale protein function classification,” (in eng),
Bioinformatics, vol. 30, no. 9, pp. 1236-40, May 2014.
- [9] S. Tang, A. Lomsadze, and M. Borodovsky, “Identification of protein coding regions in
RNA transcripts,” Nucleic Acids Research, 2015