Interpreting the Results
EnTAP provides many output files at each stage of execution to better see how the data is being managed throughout the pipeline:
Protein Families (optional)
The two files to check out first are the final annotations and log file. These files contain a summary of all the information collected at each stage, including statistical analyses. The remaining files are there for a more in depth look at each stage. All files will be contained in “entap_outfiles” directory as default, or different if the - - out-dir flag was specified.
Final Annotations
The final EnTAP annotations are contained within the /final_results directory. These files are the summation of each stage of the pipeline and contain the combined information. So these can be considered the most important files! The “full_entap.tsv” file will contain all of the information gathered throughout the pipeline summarized in one file. This will include annotated, unannotated, and contaminated sequences.
All .tsv files in this section may have the following header information (from left to right) separated by each portion of the pipeline. Some headers will not be shown if that part of the pipeline was skipped or the information was not found for any of the input sequences. TSV formatted files support Tidyverse format (including ‘NA’ being used for empty data cells).
- General Header Information
Query sequence ID
- Frame Selection Header Information (optional)
Open Reading Frame
- Expression Analysis Header Information (optional)
FPKM
TPM
Effective Length
- Similarity Search Header Information
Subject sequence ID
Percentage of identical matches
Alignment length
Number of mismatches
Number of gap openings
Start of alignment in query
End of alignment in query
Start of alignment in subject
End of alignment in subject
Expect (e) value
Query coverage
Subject title
Species
Taxonomic Lineage
Origin Database
Contaminant (yes/no if the hit was flagged as a contaminant)
Informative (yes/no if he hit was flagged as informative)
- Similarity Search UniProt Header Information (optional if aligning against SwissProt database)
UniProt Database Cross References
UniProt Additional Information
UniProt KEGG Terms
UniProt GO Biological
UniProt GO Cellular
UniProt GO Molecular
- Ontology EggNOG Header Information
Seed Ortholog
Seed E-Value
Seed Score
Predicted Gene
Taxonomic Scope
OGs (orthologous groups assigned)
EggNOG Description (EggNOG)
KEGG Terms (EggNOG)
GO Biological (Gene Ontology)
GO Cellular (Gene Ontology)
GO Molecular (Gene Ontology)
BIGG Reaction
- Ontology InterProScan Header Information
IPScan GO Biological
IPScan GO Cellular
IPScan GO Molecular
Pathways
InterPro (InterPro database entry)
Protein Database (database assigned. Ex: pfam)
Protein Description (description of database entry)
E Value (E-value of hit against protein database)
full_entap.tsv
This .tsv file is essentially a final report from EnTAP that will have the headers as mentioned previously, summarizing the results of the entire pipeline
Since this includes every single transcript, there will be annotated, unannotated, and contaminated sequences. Further filtering of transcripts (for example if you are only interested in those transcripts that were annotated) can be done with this file or the below files
annotated.faa / .fnn / .tsv
Nucleotide/protein fasta files along with tsv file containing all sequences that either align databases through similarity searching or through the ontology stage
unannotated.faa / .fnn / .tsv
Nucleotide/protein fasta files along with tsv file containing all sequences that did not align either through similarity searching nor through the ontology stage
annotated_contam.faa / .fnn / .tsv
Nucleotide/protein fasta files along with tsv file containing all annotated sequences that were flagged as a contaminant
annotated_without_contam.faa / .fnn / .tsv
Nucleotide/protein fasta files along with tsv file containing all annotated sequences that were not flagged as a contaminant
x_enrich_geneid_go.tsv
Tab-deliminated file that can be used for Gene Enrichment
First column contains the gene ID and second column contains the Gene Ontology term corresponding to the gene ID
x_enrich_geneid_len.tsv
Tab-deliminated file that can be used for Gene Enrichment
First column contains the gene ID and second columns contains the effective length from Expression Analysis. This file will not be printed if Expression Analysis has not been ran
Note: the Length column will not be printed when Expression Filtering has not been performed
x_gene_ontology_terms.tsv
Tab-deliminated file that can be used for Gene Enrichment
Columns are as follows: Sequence ID, Gene Ontology Term ID, Gene Ontology Term, Gene Ontology Category, and Effective Length
Note: the Effective Length column will not be printed when Expression Filtering has not been performed
Log File / Statistics
The log file contains a statistical analysis of each stage of the pipeline that you ran. I’ll give a brief outline of some of the stats performed:
Initial Statistics
Transcriptome statistics: n50, n90, average gene length, longest/shortest gene
Summary of user flags
Summary of execution paths (from config file)
Expression analysis
Transcriptome statistics: n50, n90, average gene length, longest/shortest gene
Summary of sequences kept/removed after filtering
Frame Selection
Transcriptome statistics: n50, n90, average gene length, longest/shortest gene
Summary of frame selection: Partial, internal, complete genes. Genes where no frame was found
Similarity Searching
Contaminant/uninformative/informative count
Phylogenetic/contaminant distribution of alignments
Alignment distribution based upon frame results (partial/internal/complete)
Sequence count that did not align against a database reference
Statistics calculated for each individual database and final results
Gene Family Assignment
Phylogenetic distribution of gene family assignments
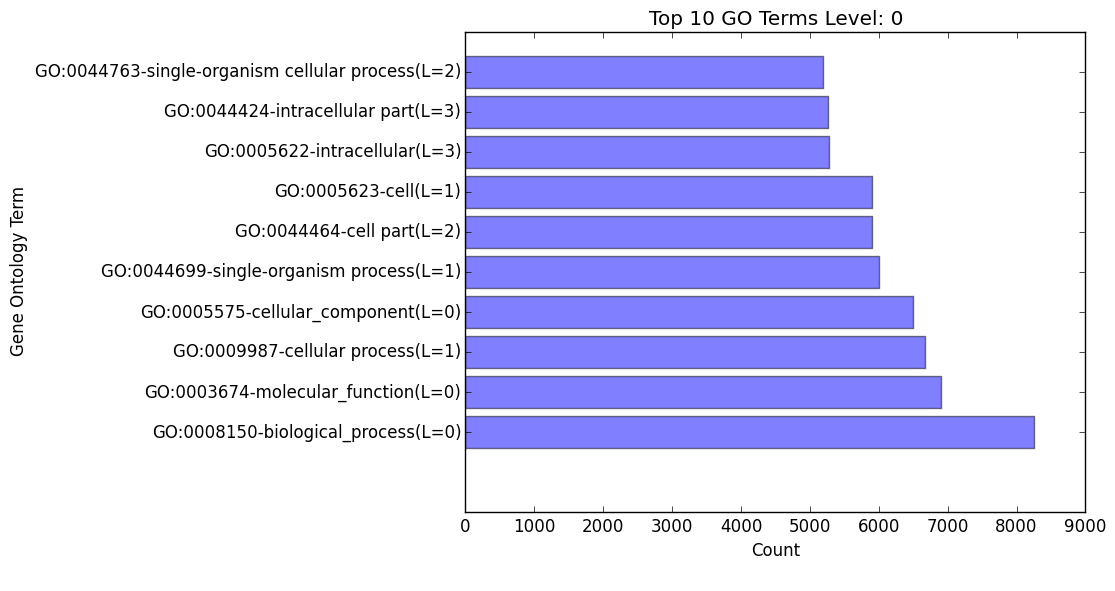
Gene Ontology category distribution (biological processes, molecular function, cellular component)
InterProScan
Additional statistics coming soon!
Final Annotation Statistics
Statistical summary of each stage
Runtime
Transcriptomes
The /transcriptomes contains the original, processed, and final transcriptomes being used by EnTAP. The files are as follows with the ‘transcriptome’ tag based upon the name of your input transcriptome:
transcriptome.fasta
This file is essentially a copy of your input transcriptome. The sequence ID’s may be changed depending on whether you selected the ‘trim’ flag or otherwise.
transcriptome_expression_filtered.fasta
As the name implies, this transcriptome is the resultant of the Expression Filtering stage with sequences removed that fall under the FPKM threshold you have specified.
transcriptome_frame_selected.fasta
This transcriptome is the resultant of Frame Selection. Sequences in which a frame was not selected are removed and those with a frame are kept in this file. As a result, this file will always be in protein format.
transcriptome_final.fasta
This is your final transcriptome following the “Transcriptome Filtering” stage of EnTAP. This transcriptome will be used for the later stages of the pipeline (Similarity Searching and Ontology). Depending on which methods of execution you chose (runN / runP), the result here may be either protein or nucleotide with Frame Selection and/or Expression Filtering.
Expression Filtering (RSEM)
The /expression folder will contain all of the relevant information for this stage of the pipeline. This folder will contain the main files (results from expression analysis software), files processed from EnTAP (including graphs).
RSEM Files: /expression
The /expression directory will contain all of the output from RSEM including a converted BAM file (if you input a SAM) and the results of the expression analysis.
EnTAP Files: /processed
This directory will contain all of the files produced from EnTAP concerning expression analysis. With a generic transcriptome input of “Species.fasta”, these files will have the following format
Species_removed.fasta
Fasta file of sequences that were under the specified FPKM threshold
Species_kept.fasta
Fasta file of sequences that were kept after filtering (over the FPKM threshold)
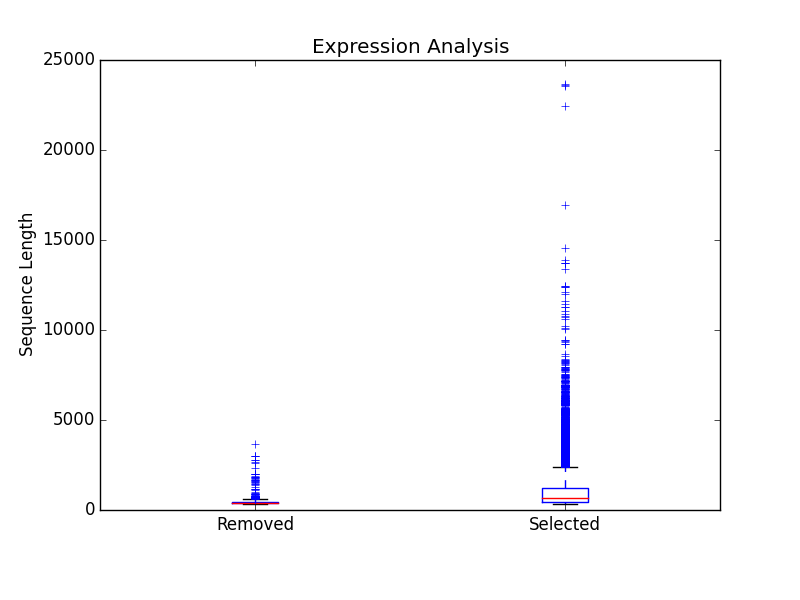
/figures
Directory containing a box plot of sequence length vs the sequences that were removed and kept after expression analysis
Frame Selection (GeneMarkS-T or TransDecoder)
The /frame_selection folder will contain all of the relevant information for the frame selection stage of the pipeline. This folder will contain results from frame selection software, files processed from EnTAP, and figures generated from EnTAP.
TransDecoder Files: /frame_selection
The files within the root /frame_selection directory contain the results from the frame selection portion of the pipeline. More information can be found at TransDecoder (the descriptions below are taken from there):
transcripts.fasta.transdecoder.pep
Peptide sequences for the final candidate ORFs; all shorter candidates within longer ORFs were removed.
transcripts.fasta.transdecoder.gff3
Positions within the target transcripts of the final selected ORFs
transcripts.fasta.transdecoder.cds
Nucleotide sequences for coding regions of the final candidate ORFs
.err and .out file
These files are will contain any error or general information produced from the TransDecoder run
GeneMarkS-T Files: /frame_selection
The files within the root /frame_selection directory contain the results from the frame selection portion of the pipeline. More information can be found at GeneMarkS-T. With a generic transcriptome input of “Species.fasta”, these files will have the following format:
Species.fasta.fnn
Nucleotide fasta formatted frame selected sequences
Species.fasta.faa
Amino acid fasta formatted frame selected sequences
Species.fasta.lst
Information on each sequence (partial/internal/complete/ORF length)
.err and .out file
These files are will contain any error or general information produced from the GeneMarkS-T run
EnTAP Files: /processed
Files within the /processed are generated by EnTAP and will contain ORF information based on the GeneMarkS-T or TransDecoder execution. Using TransDecoder as an example:
transdecoder_complete_genes.fasta
Amino acid sequences of complete genes from transcriptome
transdecoder_partial_genes.fasta
Amino acid sequences of partial (5’ and 3’) sequences
transdecoder_internal_genes.fasta
Amino acid sequences of internal sequences
transdecoder_sequences_lost.fasta
Nucleotide sequences in which a frame was not found. These will not continue to the next stages of the pipeline
EnTAP Files: /figures
In addition to files, EnTAP will generate figures within the /figures directory. These are some useful visualizations of the information provided by GeneMarkS-T
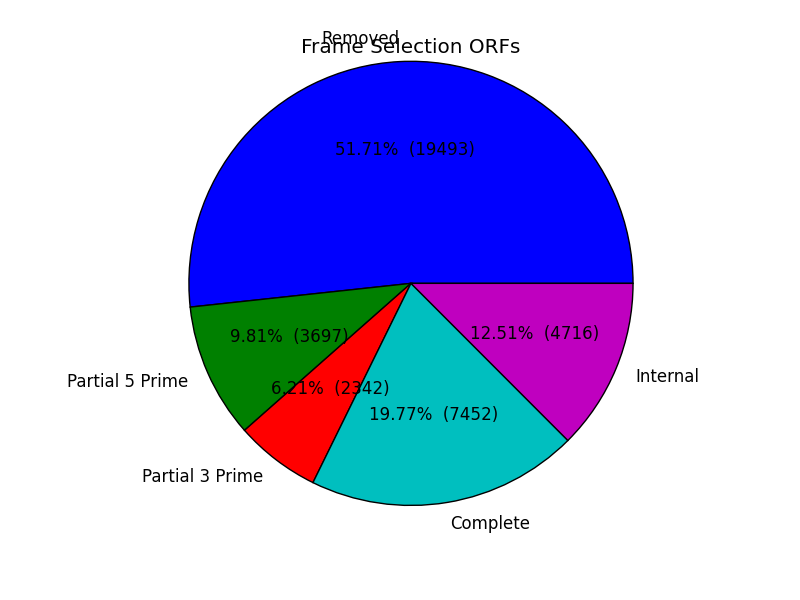
frame_results_pie.png
Pie chart representing the transcriptome (post expression filtering) showing complete/internal/partial/and sequences in which a frame was not found
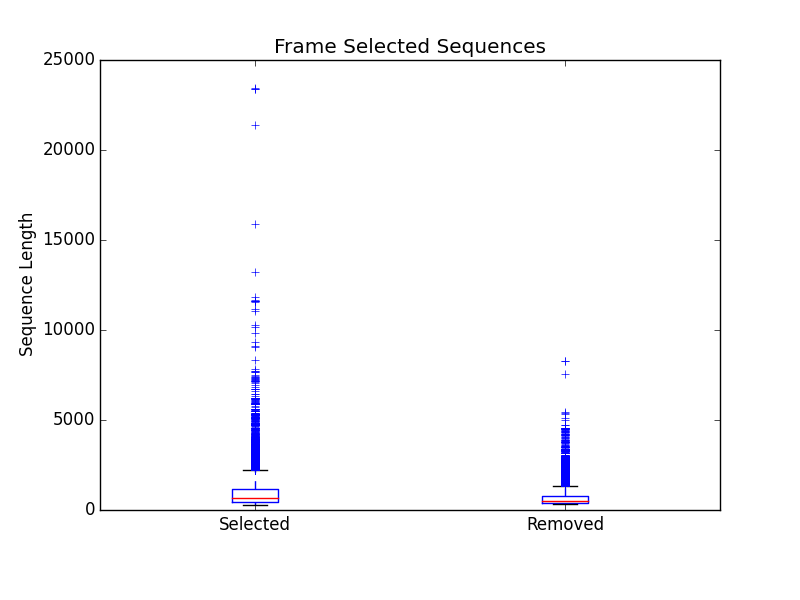
frame_selected_seq.png
Box plot of sequence length vs. the sequences that were lost during frame selection and the sequences in which a frame was found
Similarity Search (DIAMOND)
The /similarity_search directory will contain all of the relevant information for the similarity searching stage of the pipeline. This folder will contain the main files (results from similarity search software), files analyzing hits from each database, overall results combining the information from each database, and figures generated from EnTAP.
DIAMOND Files: /similarity_search
The files within the /similarity_search directory contain the results from the similarity searching portion of the pipeline against each database you select. More information can be found at DIAMOND. With running _blastp (protein similarity searching), a generic transcriptome input of “Species.fasta”, with a database called “database” the files will have the following format:
blastp_Species_database.out
This contains the similarity search information provided in the format from DIAMOND
Header information (from left to right):
Query Sequence ID
Subject Sequence ID
Percentage of Identical Matches
Alignment Length
Number of Mismatches
Number of gap openings
Start of alignment in query
End of alignment in query
Start of alignment in subject
End of alignment in subject
Expect (e) value
Bit score
Query Coverage
Subject Title (pulled from database)
blastp_Species_database_std.err and .out
These files are will contain any error or general information produced from DIAMOND
EnTAP Files: /processed
Files within the /processed are generated by EnTAP and will contain information based on the hits returned from similarity searching against each database. This information contains the best hits (discussed previously) from each database based on e-value, coverage, informativeness, phylogenetic closeness, and contaminant status.
The files below represent a run with the same parameters as the section above:
All the TSV files mentioned in this section will have the same header as follows (from left to right):
Query sequence ID
Subject sequence ID
Percentage of identical matches
Alignment length
Number of mismatches
Number of gap openings
Start of alignment in query
End of alignment in query
Start of alignment in subject
End of alignment in subject
Expect (e) value
Query coverage
Subject title
Species (pulled from hit)
Origin Database
ORF (taken from frame selection stage)
Contaminant (yes/no the hit was flagged as a contaminant)
database/diamond_annotated.faa and .fnn and .tsv
Best hits (protein and nucleotide) that were selected from this database
This contains ALL best hits, including any contaminants that were found as well as uninformative hits. Sometimes a contaminant can be the highest quality alignment!
The .tsv file contains the header information mentioned above of these same sequences
Note: Protein or nucleotide information may not be available to report depending on your type of run (these files will be empty)
database/diamond_annotated_contam.faa/.fnn/.tsv
Contaminants (protein/nucleotide) separated from the best hits file. As such, these contaminants will also be in the _best_hits.faa/.fnn.tsv files
database/diamond_annotated_without_contam.faa/.fnn/.tsv
Sequences (protein/nucleotide) that were selected as best hits and not flagged as contaminants
With this in mind: best_hits = best_hits_no_contam + best_hits_contam
These sequences are separated from the rest for convenience if you would like to examine them differently
database/unannotated.faa/.fnn/.tsv
Sequences (protein/nucleotide) from the transcriptome that did not hit against this particular database.
This does not include sequences that were lost during expression filtering or frame selection
database/diamond_unselected_hits.tsv
Similarity searching can result in several hits for each query sequence. With only one best hit being selected, the rest are unselected and end up here
Unselected hits can be due to a low e-value, coverage, or other properties EnTAP takes into account when selecting hits
EnTAP Files: /overall_results
While the /processed directory contains the best hit information from each database, the /overall_results directory contains the overall best hits combining the hits from each database.
EnTAP Files: /figures
In addition to files, EnTAP will generate figures within the /figures directory for each database. These are some useful visualizations of the information provided by similarity searching.
Here, there will be several figures:
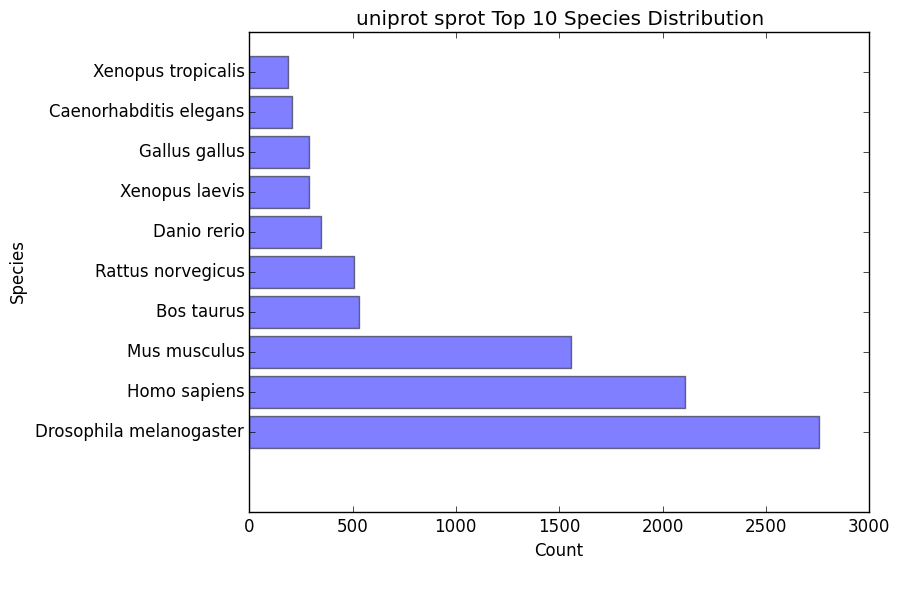
species_bar.png / species_bar.txt
Bar graph representing the top 10 species that were hit within a database
Text file representing the data being displayed
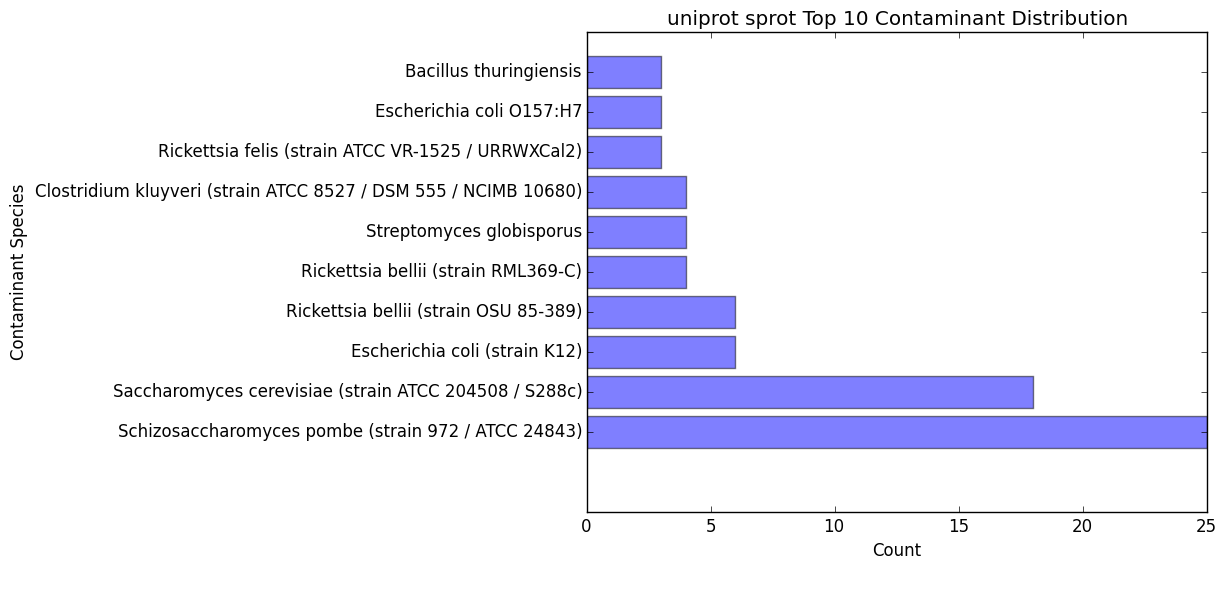
contam_bar.png / contam_bar.txt
Bar graph representing the top 10 contaminants (within best hits) that were hit against the databast
Text file representing the data being displayed
Orthologous Groups/Ontology (EggNOG)
The /ontology/EggNOG directory will contain all of the relevant information for the EggNOG stage of the pipeline. This folder will contain the EggNOG files, files analyzing the annotation from EggNOG, and figures generated from EnTAP.
EggNOG Files: /ontology/EggNOG
Files within the /ontology/EggNOG are generated through DIAMOND alignment against the EggNOG orthologous database and will contain information based on the hits returned. More information can be found at EggNOG.
blastp_transcriptome_eggnog_proteins.out
EggNOG results for sequences from the final transcriptome being used (post-processing)
EnTAP Files: /processed
Files within the /processed are generated by EnTAP and contain information on what sequences were annotated and which were not.
eggnog_unannotated.fnn/faa
Sequences where no gene family could be assigned (nucleotide/protein)
eggnog_annotated.fnn/faa
Sequences where a gene family could be assigned (nucleotide/protein)
EnTAP Files: /figures
The /figures will contain figures generated by EnTAP of Gene Ontology and Taxonomic distribution of the results
(overall/molecular_function/cellular_component/biological_process)#_go_bar_graph.png/.txt
Bar graph of each category of Gene Ontology terms
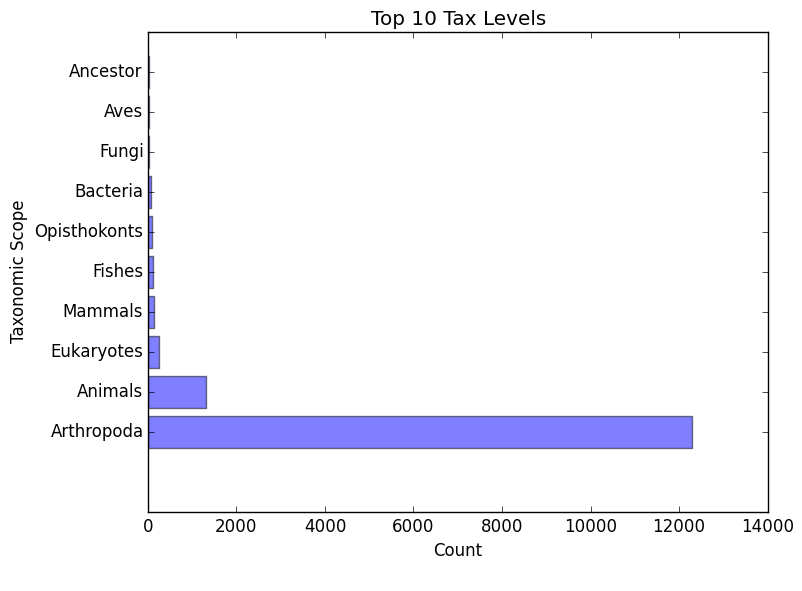
eggnog_tax_scope.png/.txt
A bar graph representation of the taxonomic scope of the gene families assigned through EggNOG
Protein Families (InterProScan)
The /ontology/InterProScan directory will contain all of the relevant information for the optional InterProScan stage of the pipeline. This folder will contain the InterProScan files and files generated from EnTAP analyzing the annotation from InterProScan.
InterProScan Files: /ontology/InterProScan
Files within the /ontology/InterProScan are generated through InterProScan and will contain information based on the results from the InterPro databases. More information can be found at InterProScan.
interproscan.tsv/xml
Tab delimited or XML file containing information on the sequences with domain matches. Information such as signature accession/description information and GO/Pathway alignments.
EnTAP Files: /processed
Files within the /processed are generated by EnTAP and contain information on what sequences had domain matches, and which did not.
unannotated_sequences.fnn/faa
Sequences where no domain could be assigned (nucleotide/protein)
annotated_sequences.fnn/faa
Sequences where a no domain could be assigned (nucleotide/protein)